Abstract
Purpose
The viscosity of highly concentrated therapeutic monoclonal antibody (mAb) formulations at concentrations ≥ 100 mg/mL can significantly affect the stability, processing, and drug product development for subcutaneous delivery. An early identification of a viscosity prone mAb during candidate selection stages are often beneficial for downstream processes. Higher order structure of mAbs may often dictate their viscosity behavior at high concentration. Thus it is beneficial to gauge or rank-order their viscosity behavior using noninvasive structural fingerprinting methods and to potentially screen for suitable viscosity lowering excipients.
Methods
In this study, Dynamic Light Scattering (DLS) and 2D NMR based methyl fingerprinting were used to correlate viscosity behavior of a set of Pfizer mAbs. The viscosities of mAbs were determined. Respective Fab and Fc domains were generated for studies.
Result
Methyl fingerprinting of intact mAbs allows for differentiation of viscosity prone mAbs from well behaved ones even at 30–40 mg/ml, where bulk viscosity of the solutions are near identical. For viscosity prone mAbs, peak broadening and or distinct chemical shift changes were noted in intact and fragment fingerprints, unlike the well-behaved mAbs, indicative of protein protein interactions (PPI).
Conclusion
Fab-Fab or Fab-Fc interactions may lead to formation of protein networks at high concentration. The early transients to these network formation may be manifested through peak broadening or peak shift in the 2D NMR spectrum of mAb/mAb fragments. Such insights go beyond rank ordering mAbs based on viscosity behavior, which can be obtained by other methods as well..
Similar content being viewed by others
Introduction
Therapeutic monoclonal antibodies (mAbs) are one of the fastest growing classes of biologics-based drugs over the past few years due to their efficacy, specificity, and potency (1,2,3). As of 2020, the commercial pipeline for mAbs is robust with around 580 + candidates in clinical trials and as many as 12 + mAbs have been approved by the FDA (Food and Drug Administration) in 2019–2020. They span across disease areas such as various forms of cancer (targeted therapy and immunotherapy), autoimmune, and respiratory disorders (2). Infact, few of them have been successfully used as treatment of SARS-COV-2 infection. Often, high-concentration mAb dosages suitable for subcutaneous injection are recommended for convenience of self-administration, enhanced medication adherence, and reduced healthcare costs (4, 5). The development of high-concentration mAbs can pose significant challenges to product development pertaining to stability, manufacturability, and processing. High-concentration mAb formulation in a prefilled syringe are well documented; however increased injection force for syringes with high fluid viscosity along withformulation stability is particularly challenging (6).
The molecular basis of elevated viscosity at high concentrations of mAbs have been extensively studied in the literature (7,8,9,10). It is well established that reversible protein–protein association at high concentrations may lead to increased solution viscosities (9, 11). Antibodies when treated as rigid charged colloidal particles, exhibit self-association propensities and elevated viscosities, at pH closer to pI (12, 13). Thus, at pH further away from pI, presence of a net charge results in charge-charge repulsion, which lowers viscosity. This may not explain the high viscosity of mAbs formulated at pH further away from their pI (13, 14). Heterogeneous surface charge distribution on the mAb can lead to attractive electrostatic interactions that cause increased viscosity build even when formulated away from the mAb pI. In addition, electrostatic interactions are not the sole determinant of the PPI leading to increased solution viscosities (11, 15,16,17). Indeed, hydrophobic interactions across Fab-Fab, Fab-Fc domains are also responsible for increased solution viscosities of high concentration mAbs (18, 19). The complex nature of PPI in high concentration mAb solutions has been qualitatively predicted from either interaction parameter (kD) from dynamic light scattering (DLS) methods or from affinity-capture self-interaction nanoparticle spectroscopy (AC-SINS), measured under very dilute concentrations (20,21,22,23). Additionally, several methods like neutron scattering, static light scattering, and X-ray scattering have been employed to probe the nature of PPI in high concentration mAb formulations (7, 8, 10, 24,25,26,27). However, extrapolation of protein solution behavior at dilute conditions to behavior at high concentration may not be straightforward (28). The nature of PPI leading to higher order species may vary significantly with changes in concentrations (26, 29,30,31). Furthermore, incompatibility of formulation buffer components can also interfere with these biophysical assays, which makes interpretation of the results challenging (21, 32). The techniques widely used for probing the mechanism of viscosity are mostly low resolution and offer little or no information of the inherent structural, conformational, or underlying molecular basis that lead to an understanding of the behavior of mAbs at high concentrations. Such high-resolution information at a domain or peptide level is strongly desired across various stages of drug product development and are useful to (a) identify poorly behaved mAbs in the early developmental stages, (b) identify and engineer molecular determinants of viscosity, and/or (c) rational design of viscosity lowering agents for a given mAb (33,34,35,36,37,38). Specifically, the choice of viscosity lowering excipients can vary for different mAbs depending upon the primary sequence and surface exposure of specific amino acids of Fab (fragment antigen-binding) domains even under similar solution conditions implying PPI may be unique for each mAb (16, 34, 39,40,41,42,43). The contribution of individual domains of mAb (i.e. Fab-Fab interactions and Fab-Fc interactions) in mediating high viscosity has been demonstrated in two different case studies (12, 44). Recently, hydrogen–deuterium exchange mass spectrometry has enabled better characterization of domain specific interactions of mAb, which have shed light on the intricacies of PPI in mediating viscosity at the peptide level (45, 46).
In this work, we aim to probe the role of PPI in mediating viscosity of therapeutic mAbs at an atomistic level. A set of mAbs, having similar pI, but varying viscosity under identical buffer conditions, were selected for this study. The nature of PPI in these intact mAbs were probed using DLS-derived interaction parameters (kD). Higher order structure (HOS) fingerprints of intact mAbs and respective fragments were obtained by high resolution 2D [1H- 13C] methyl NMR methods to identify viscosity prone mAbs. The impact of viscosity lowering agents on the HOS fingerprints of mAbs were also characterized by 2D NMR. Finally, using 2D NMR fingerprints acquired at moderate protein concentrations in the presence and absence of viscosity-reducing agents, were used to validate the robustness of methodology.
Materials and Methods
All materials used in the study are reagent grade or better. Amicon concentrators and filters were obtained from Millipore (Billerica, Massachusetts). Glass vials were obtained from West Pharmaceutical Services (Exton, Pennsylvania). FragIT—Immobilized FabRICATOR (IdeS) enzyme on agarose spin columns, were purchased from Genovis Inc. NMR tubes were obtained from Sigma Aldrich and Bruker Biospin. For NMR, 99% D2O was obtained from Sigma Aldrich (St. Louis).
mAbs Used in this Study
The therapeutic monoclonal antibodies (mAb) used in the study are described in Table S1.The experimental workflow has been guided by (a) the need to test the applicability of the method across few mAbs across isotypes and (b) sample availability. Based on the viscosity values in Fig. 1, the mAbs can be ordered as mAb-1 < < mAb-2 < mAb-3 < mAb 4 < mAb-5. The isoelectric point (pI) of all the mAbs are similar (6.8–7.1). mAb-1, mAb-3, and mAb-5 corresponds to IgG1 isotype and mAb-2 and mAb-4 corresponds to IgG2 subtype. mAb-1 (control mAb) and mAb-2 serves as well-behaved control for IgG1 and IgG2 isotypes, respectively.
Sample Preparation
All the mAbs and mAb fragments were dialyzed into 20 mM Histidine at pH 5.5. For NMR experiments, the concentration of intact mAbs were ~ 30–40 mg/mL. Fragments of mAb 1 and 5 were generated by digesting intact mAbs using Fabricator (IdeS). IdeS is a cysteine protease that cleaves monoclonal antibodies at the -GPSV sequence just below the hinge region to generate Fc and Fab2 domains. For each of the digestion reaction, 30–50 mgs of mAbs were used. The digestion was performed between 18 and 24 h at ambient temperature. Capture Select Maxi Spin columns were used for purification and vendor’s protocol was followed with minor modifications. Fab2 and Fc were separately dialyzed against 2 L of 20 mM histidine at pH 5.5. For mAb-2 and mAb-5, digestion was performed at a protein concentration of 36 mg/mL and the digested mixture was briefly centrifuged and subsequently dialyzed into 20 mM Histidine pH 5.5. Final concentration of digested fragments was approximately 15–20 mg/mL.
Dynamic Light Scattering
For Dynamic Light Scattering (DLS) studies, appropriate dilutions in the range of 2–10 mg/mL were prepared in the same formulation buffer (20 mM histidine/histidine-HCl buffer, pH 5.5). DLS was performed using a Dynapro Plate Reader II (Wyatt Technology) and analyzed using Dynamics V7.1.7 software. 35 μL of sample was added to a 384 Well UV-StarÒClear Microplate (781801) (Greiner Bio-One GmbH, Germany). The plate was covered with a clear disposable tape, Topseal™ (Packard – Meriden,CT), to avoid evaporation during measurement. Prior to measurement all plates were centrifuged at 3000 rpm for 5 min using an Eppendorf Centrifuge 5810 R (Eppendorf AG, Hamburg). 10 acquisitions of each 5 s were collected for each sample with auto adjustment of attenuator and laser power. Three wells of each sample were averaged, and all samples were measured in triplicate. Water was chosen as the default solvent assuming no significant changes in viscosity in the 2–10 mg/mL protein concentration range. The mutual diffusion coefficient, Dm (m2/s) of the mAbs was used to obtain the protein–protein interaction parameter, kD, using the equation below. Ds is the self-diffusion coefficient (the value of Dm at infinite dilution) and c is the protein concentration. Ds is obtained from the intercept of a plot of Dm versus C and kD can be obtained from the slope of the curve.
Viscosity Measurement
All viscosity measurements were carried out at 25 ºC using m-VROC Viscometer (RheoSense, San Ramon, CA). or Anton Paar CP25-1 cone and plate rheometer method as described in previous reports (47). Briefly, mAb samples were concentrated to their solubility limits in 20 mM Histidine, pH 5.5 and then a serial dilution was performed prior to viscosity measurements. All viscosity measurements were performed between shear rates of 500–10,000 (1/s).Viscosity data at 100 mg/mL mAb concentration were used for the purpose of assessing correlations between high concentration viscosity and higher order structural changes from NMR. In order to gauge the impact of increased solution viscosity on NMR fingerprints of a mAb, two samples were made (a) 30 mg/mL mAb2 and (b) 30 mg/ml mAb2 and 85% (w/v) glycerol. The viscosity of the two samples were measured and 2D NMR of the two samples were taken.
Nuclear Magnetic Resonance Spectroscopy
The concentration of intact mAbs and mAb fragments have been previously optimized for Nuclear Magnetic Resonance (NMR) studies (48, 49). 500 μL of sample were used for NMR analysis.The Fc and Fab2 concentrations used for NMR experiments were ~ 15 to 20 mg/mL. In general, for the intact mAbs, increased viscosity results in poor Signal/Noise (S/N) for [1H-13C] Heteronuclear Single Quantum Coherence Spectroscopy (HSQC) experiments acquired at 50 mg/ml sample and 50 °C (data not shown) (50). The intact mAbs were used at a concentration of 30–40 mg/mL. All the NMR samples had 10% (by volume) D2O. Unless otherwise noted, all the NMR spectra were acquired on a 900 MHz Bruker Avance III spectrometer equipped with triple resonance cryogenically cooled TCI probes (proton-optimized triple resonance NMR 'inverse' probe) with a single axis gradient system. 13C-methyl fingerprint data were collected at 50 C. 2D [1H-13C] HSQC (sensitivity enhanced, echo-antiecho) spectra were recorded with 280 scans per increment, a recycle delay of 1 s, 2048 × 128 complex points for a spectral width of 15 ppm by 18 ppm in the proton, and carbon dimension, respectively. NMR data was processed and analyzed using Topspin 3.5. The entire procedure of fragmentation, purification and NMR data acquisition for mAb-1, mAb-2 and mAb-5 could be performed, but not for mAb-3 and mAb-4 due to limiting sample availability. Hence a comparison of 2D NMR spectra of intact mAb and digestion mixture was performed for mAb-3 and mAb-4. Enzyme digested mixtures were used instead of purified Fab2 and Fc for these two mAbs.
Results
A set of mAbs with varying viscosities were selected for this study. Biophysical parameters such as protein–protein interaction parameter (kD) and NMR derived higher order structure fingerprint were used to identify viscosity prone mAbs. The HOS fingerprints were used to identify the molecular basis of solution viscosities and the ability of certain solution conditions to lower their viscosities.
Interaction Parameters and Viscosity Assessment of mAbs
To closely compare the molecular properties and to probe the structural determinants responsible for viscosity behavior, a panel of 2 well behaved mAbs and 3 poorly behaved mAbs were assembled with a wide range of viscosities. The selected samples were further classified based on the isotype class (IgG1 vs IgG2) as shown in Table S1. In the following discussion, mAb-3 and mAb-5 will be compared to the mAb-1; mAb-4 will be compared to mAb 2.
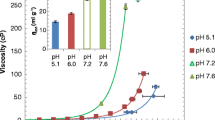
Figure 1A shows the viscosity profile of all the selected mAbs. At 100 mg/mL, mAb-5 has the highest viscosity followed by mAb-4 and mAb-3. The control/well-behaved mAbs, mAb-1 and mAb-2, are well within the acceptable viscosity of 20 cP,. Additionally, when glycerol was added to mAb-2 solution by 85% w/v, the viscosity increased to 66 cP (Table S1). Increase in viscosity can be attributed to increased molecular crowding and excluded volume effects in mAb solutions due to solution properties of glycerol.
DLS-derived kD measurements were performed to identify colloidal behavior of mAbs 1–5 samples at dilute concentration ranges (2–15 mg/mL) using linear fits of mutual diffusion coefficient (Dm) plots. Several studies have been used in earlier reports, to interpret kD in terms of attractive or repulsive PPI. The net PPI is neutral around -6 to -9 mL/g and kD values lower than this cutoff has been interpreted to be attractive PPI (24). Since the absolute values of kD may vary according to the buffers and salt conditions, the mAbs in this study were all studied under identical buffer condition. The results from the DLS experiments are presented in Fig. 1B and in Table S1. Across the mAbs, only mAb-1 has a positive kD, suggestive of net repulsive protein protein interactions. All the viscosity prone mAbs mAb-3, 4, 5 have increasingly negative kD. The well behaved mAb-2 has kD -8.7 mL/g which indicates no net PPI. Based on the kD, values, it appears that for the viscosity prone mAbs, net self-association interactions are present even in dilute concentration regime which could be amplified at higher protein concentrations (> 100 mg/mL). Last, but not the least, Do value for mAb -5 suggests the formation of oligomeric species of mAb-5 even under infinite dilution, unlike any other mAbs used in the study.
Nuclear Magnetic Resonance Spectroscopy
The chemical shifts and peak intensities of methyl group in 2D [1H- 13C] HSQC spectrum are diagnostic of respective higher order structure (HOS) of mAbs. The methyl group bearing residues (Ile, Leu, Val, Thr, Met, Ala) are generally distributed uniformly throughout mAb sequence and are also important markers for protein–protein interactions. Contrary to backbone methyl amides, they are less exposed and hence are less sensitive to solvent exchanges mediated by change in solution conditions (pH etc.) (51). To this end HOS fingerprints of a mAb in a dilute solution and a highly viscous solution needs to be compared. Compared to a dilute solution, a highly viscous solution represents a crowded environment for proteins. Under these conditions, protein molecules are located closer to one another and exhibits protein–protein interactions which may be otherwise absent in dilute solutions. These interactions ensure that the protein is no longer an isolated molecule but is part of interacting network of molecules. As a result, rotational tumbling of the protein decreases and transverse relaxation rate (T2 = 1/R2) increases. This is manifested by homogeneous line broadening in the 2D [1H- 13C] HSQC spectrum. In order to demonstrate the impact of high solution viscosity on the NMR fingerprints, 2D [1H- 13C] HSQC of a well behaved control mAb-2 were taken under (a) dilute buffer condition and (b) mAb-2 + 85% by wt glycerol (Fig. S1). As is evident from the spectra, there is less number of peaks in methyl region of the spectra (homogeneous peak broadening) in glycerol environment, which implies non-specific protein–protein interaction. It is important to note that glycerol induced non-specific protein–protein interaction may not be identical to specific PPI, if any, that can also lead to increased solution viscosity in high concentration mAb solutions. Since mAb-5 (IgG1) has highest viscosity compared to mAb-1 at 100 mg/mL, the HOS fingerprints of mAb-1 and mAb-5 were compared using 2D NMR. The NMR experiments for the two mAbs were performed at similar concentrations (30–40 mg/ml) where viscosity values are comparable (Fig. 1A). Surprisingly, 2D [1H- 13C] HSQC spectra was able to differentiate between mAb-1 and mAb-5. While mAb-1 shows a well dispersed [1H-13C] spectrum, almost all the cross peaks of mAb-5 were broadened beyond detection (Fig. 2A). Given the viscosity values of the two mAbs being similar at this concentration, observed peak broadening can be potentially ascribed to intermolecular Fab-Fab, Fab-Fc or Fc-Fc interactions. To further investigate the role of individual domains (Fab-2 or Fc) responsible for PPI, the HOS fingerprints of independent Fab2 and Fc domains were also acquired. The 2D [1H- 13C] HSQC spectra of Fc domain of mAb-5 and mAb-1 are nearly identical with comparable S/N (Fig. 2B). This clearly rules out any Fc-Fc interaction that might cause spectral broadening in the intact mAb-5. On the other hand, the Fab-2 domain of mAb-1 and mAb-5 shows contrasting results. While Fab-2 of mAb-1 yields a well dispersed 2D [1H- 13C] spectra, cross peaks for mAb-5 in the methyl region (2.0 ppm to -1.0 ppm) were homogeneously broadened beyond detection This suggests inter-molecular interaction between Fab domains are also present in Fab2 fragment. Since IdeS cleavage did not perturb the overall Fab2 domain architecture, such Fab2-Fab2 interactions that are present in intact mAbs are also retained in the cleaved Fab2 fragments (Fig. 2C). Thus for mAb-5, the disappearance of peaks compared to control mAbs results in poor S/N ratio and is indicative of non-specific protein–protein association. This type of association can, in principle, be reconstituted even in dilute control mAb solution (mAb-2), by adding 85% glycerol.
Impact of protein–protein association on HOS fingerprints of intact mAb (A), Fc (B) and Fab2 (C) domain for a well-behaved IgG1 mAb (mAb-1) and viscosity-prone IgG1 mAb (mAb-5). For mAb-5, [1H-13C] HSQC spectra of intact mAb and Fab2 does not yield any cross peaks when compared to those of mAb-1. The [1H-13C] HSQC spectra of Fc fragment for mAb-1 and mAb-5 are similar with respect to number of peaks and peak disposition.
Since increased protein–protein association results in differential line broadening for mAb-5, any excipient that inhibit protein–protein association should also increase S/N of the 2D [1H- 13C] HSQC spectra. Indeed, comparison of intact mAb-5 in 100 mM arginine buffer at pH 5.5 shows moderately improved spectral quality as evidenced by the appearance of peaks in Fig. 3A. Similar effect is also observed for Fab-2 of mAb-5 in the presence of 100 mM arginine at pH 5.5 (Fig. 3B). Based on the inspection of the spectra, it appears that arginine can disrupt protein–protein association resulting in better S/N and appearance of more number of peaks in the methyl region of the spectrum. To further support the role of arginine, viscosity of mAb-5 was measured in the presence of arginine at pH 5.5 and pH 7.0. As shown in Fig. S2, disruption of protein–protein association by arginine is more effective at pH 7.0, which is also closer to pI of the molecule. It is important to note that at 30 mg/ml concentration, both mAb-1 and mAb-5 are indistinguishable based on their viscosity values, but clearly distinguishable in the HOS fingerprints.
The validity of the approach was tested for another viscosity prone mAb, mAb-4, along with a less viscous mAb-2 of the same IgG2 isotype. Despite being prone to viscosity, cross peaks for mAb-4 mAb were visible in in 2D [1H-13C] HSQC spectra These peaks were not homogeneously broadened, unlike mAb-5 (Fig. 4). In the methyl region, especially in the Leucine/Valine region of the spectra more peaks in mAb-2 were visible compared to that of mAb-4. Compared to mAb-2, however, the 2D [1H- 13C] spectra of mAb-4 shows significant differential peak broadening. It is important to note that upon digestion, mAb-4 spectrum shows more peaks compared to that of intact mAbs, which is expected as the molecular weight diminishes from 150 to 100 kDa and 50 kDa. This suggests that digestion has indeed happened. The digestion method have been already optimized for the system of mAbs. Careful inspection of the spectra of mAb-2, mAb-4 and mAb-5 Fc suggests that majority of the cross peaks in mAb-2 and mAb-4 belong to their respective Fc domain. The highlighted cross peak of mAb-4 belonging to Fc fragment nearly overlays with that of mAb-2, suggesting that enzymatic cleavage may have disrupted some Fc-Fab interaction in the mAb-4. The role of Fab-2–Fab2 interactions in differential broadening of mAb-4 spectra could not be ascertained from the unpurified digestion mixture. Moreover, IgG2 Fabs are also prone to disulfide scrambling between isoforms which can also lead to differential broadening, regardless of any viscosity effects (52). While the precise nature of PPI in the case of mAb-4 may not be known based on the HOS fingerprints, it suggests the presence of intermolecular Fab-Fc interaction. At least two peaks in the HSQC spectra of intact mAb-4 show chemical shift changes compared to that of mAb-2 (Fig. 4A). Enzymatic digestion alters Fab-Fc interaction and hence results in specific changes in these peaks (Fig. 4B). These peaks belong to Fc fragment of mAb-2 as highlighted in Fig. 4.
Impact of self-association in mAb-2 and mAb-4 (A) [1H-13C] HSQC spectra of intact mAb-2 yields more peaks than intact mAb-4, suggestive of the presence of protein–protein association in mAb-4 relative to mAb-2. Moreover, two peaks are highlighted for mAb-4 and mAb-2 which shows distinct chemical shift changes. One of these peaks belong to Fc domain of the two mAbs, as these peaks are visible in Fc spectra of mAb-1 as indicated by the straight line. The other peak belongs to Fab2 domain, since this is absent in Fc fragment of mAb-1. (B) Comparison of the [1H-13C] HSQC spectra of intact mAb-4 and digested mAb-4. The marked peak shows distinction chemical changes due to enzymatic digestion, potentially disrupting intermolecular Fab-Fc that exists in intact mAb.
The impact of PPI in the 2D [1H-13C] spectra of viscosity prone mAbs were further gauged by addition of viscosity lowering excipients to mAb-3 (IgG1 isotype). The kD values suggests the presence of attractive PPI in mAb-3. Unlike mAb-5 (IgG1 isotype), mAb-3 shows a well dispersed spectrum. In this regard, the viscosity for mAb-3 seems to be driven by specific Fab-Fc interactions, like mAb-2 (IgG2 isotype). In fact, selected cross peaks in the 2D [1H-13C] spectra of mAb-3 in the absence of sodium chloride nearly overlays with that of high viscosity mAb-5 as shown in Fig. S4. Addition of sodium chloride results in selected chemical shift changes, suggesting modulation of specific Fab-Fc PPI. While the two spectra are nearly superimposable, addition of salt (100 mM NaCl) induces distinct chemical shift changes in the spectra as shown in Fig. 5. For analyzing the 2D NMR data, visual comparison of the spectra have been done without taking any recourse to multivariate analysis (50).
Effect of addition of 100 mM NaCl on the HOS of mAb-3. Comparison of selected residues in mAb-3 with and without NaCl. The peaks that are labeled (square and circle) show distinct chemical shift changes, which suggests that the corresponding residues are involved in PPI. Addition of NaCl disrupts these interactions.
Similar visual comparisons were also obtained in HOS changes of Fc domain due to deglycosyaltion in case of NISTmAb, which was also evident in the 1st (dominant) mode of principle component analysis. In case of mAb-5, poor S/N renders the statistical analysis ineffective.
Discussion
Molecular Basis of Viscosity
It has been determined in scientific literature that increased solution viscosity of mAbs at high concentrations (> 100 mg/mL) may arise from reversible self-association. Such association may result in dimer formation at dilute concentrations and may progressively result in formation of larger clusters, as have been detected for high concentration monoclonal antibody solutions (9, 12) (9, 12). In other cases, formation of long lived “dynamic clusters” that may result in increasing solution viscosities (55). While the underlying mechanism(s) of viscosity behavior at a molecular level remains elusive, several biophysical techniques have been used to study PPI. The interaction parameter (kD), osmotic second virial coefficient (B22) derived from DLS, and AUC (analytical ultracentrifugation) or SLS (static light scattering), respectively, are often found to correlate with self-associative tendencies of mAb (26,27,28). To this end, for the system of mAbs that are studied here, correlation of kD and viscosity for IgG1 and IgG2 mAbs were also observed (Fig. 1C). Mechanistically, reversible self-association could be mediated by long-range electrostatic interactions which, in the absence of salts, are screened weakly (26,27,28, 32). The latter exhibits the rigid spherical colloidal nature of the proteins. While non-specific interaction could be guided by the overall charge of the mAb, specific PPI is determined by both the primary sequence and the HOS of mAb domains. The use of 2D NMR based methods to probe these specific PPI at high concentration (> 100 mg/ml) is prohibitive due to lack of sensitivity and high transverse relaxation (50), However, early onset of formation of these clusters in an intermediate concentration regime (30–40 mg/ml) is manifested in NMR fingerprint spectra either (a) from differential broadening of cross peaks or (b) specific chemical shift changes or differential broadening for a subset of peaks, when compared with control mAbs.
Higher Order Structure Fingerprinting of Monoclonal Antibodies: Orthogonal Approach to Identify a Viscosity Prone mAbs
Methyl based 2D-NMR fingerprinting of Higher Order Structure(s) of mAbs has recently been applied to probe analytical similarity, conformational dynamics, and to characterize post-translational modifications (48,49,50, 53). In the panel of mAbs used in this study, the underlying mechanism which leads to elevated viscosity of mAb-4 is quite unique. First, the number of peaks in [1H-13C] HSQC spectra is relatively more in mAb-4 compared to mAb-2, under identical conditions. Second, distinct chemical shift changes in cross peaks of mAb-4 are observed with respect to mAb-2, and as illustrated in Fig. 4A. These peaks belong to Fc domain of mAb-2 and mAb-4, as evident from the comparison of same peaks in [1H-13C] HSQC Fc of mAb-5 (Fig. 4). Upon enzymatic digestion, these peaks show distinct chemical shift in the case of mAb-4. This may imply that Fc domain in mAb-4 is possibly involved in intermolecular Fab-Fc interaction, which is lost upon enzymatic digestion (Fig. 4B). Such Fab-Fc interaction can also drive the viscosity behavior of mAb-3. For mAb-5, non-specific Fab-Fab interaction is the dominant contributor to elevated viscosity.
Our results may also indicate subtle differences in timescale of protein–protein network formation. For mAb-5, the monomeric species is in the reversible exchange with clusters/network in intermediate time scales which results in near complete broadening of the spectra. For mAb-3 and mAb-4, chemical shift changes, indicative of specific PPIs also suggest that the monomeric protein is in reversible exchange with the cluster at fast exchange time scales. In contrast control mAb-2 is devoid of any such exchange (and hence line broadening) which indicates no cluster formation. It is important to emphasize that such signatures of cluster formation is only an early transient to the overall complex network formation that may be present in high concentration. Thus, methyl based 2D fingerprinting affords (a) unprecedented resolution to identify residues (not assigned) that are involved in specific PPI and (b) qualitative timescales of cluster formation, which is strongly desired to probe molecular origins of high viscosity behavior. Specific peak broadening, as shown by 2D methyl NMR has been used to detect specific protein–protein interactions, that can lead to dimer leading to protein-aggregation as in case of Fc-fusion protein (54). However the rate of aggregation for the mAbs at 400 C do not correlate with the viscosity of the mAbs (Table S1). For example mAb-5 being the highest viscous molecule exhibits lowest aggregation. This suggests that differential peak broadening or homogeneous peak broadening are probably indicative of PPI leading to viscosity and not of aggregation.
Ability to Screen PPIs as Assessed by HOS Fingerprints
Within the premise that either Fab-Fab or Fab-Fc interactions may ultimately lead to higher viscosity at high concentration, we looked at altering solution conditions that may modulate these interactions. It is imperative to note that any excipient that disrupts the PPI could potentially serve as a viscosity lowering agent for mAbs. Indeed, 100 mM arginine screens PPI and possibly breaks up some of the “clusters” of mAb-5 in solution. The function of arginine with its charged guanidium group enables it to interact with both hydrophobic and charged surface of mAb-5. The effect of Arginine on the spectrum of mAb-5 is more prominent at pH 7.2 as it lowers the viscosity of the molecule even further (Fig. S3). Indeed, the viscosity of mAb-5 in the presence of Arginine at pH 7.2 is lower than that of pH 5.5. Since the pI of the molecule is around 7.2, it is possible that the mAb has no net charge and Arginine can screen the PPI more effectively, thereby lowering the viscosity. Alternatively, arginine can slow down the exchange between monomer and cluster, such that the S/N of mAb-5 spectra improves. On the other hand, in case of mAb-3, addition of salt impacts a definite set of peaks which likely belong to leucine/valine residues of the Fab domain (Fig. S4). Addition of salt induces distinct chemical shift changes in these peaks for mAb-3. In other words, addition of salt modulates specific Fab-Fc interaction, which is mediated by these residues. The changes in 2D NMR fingerprints for two viscosity-prone mAbs, mAb-5 and mAb-3, in the presence of viscosity lowering agents demonstrates the robustness of the method in the identification of viscosity-prone mAbs.
Limitation
One of the inherent limitation of the method is its low throughput nature, which requires sufficient amount of sample. Other methods (rheology based for example) can be used to determine viscosity of mAb solution is a faster and accurate method compared to 2D NMR. Moreover, high throughput biophysical methods like Differential Scanning Calorimetry, DLS can be used to gauge conformational stability or colloidal stability that may be related to increased solution viscosity of mAbs in some cases. However each of these methods may provide a macroscopic basis of viscosity reliably. The mechanistic basis of viscosity from residue specific or domain specific level remains elusive in this method. In this regard, 1H NMR based T2 methods have been previously used to identify viscosity of mAbs at high concentration. However, increased viscosity causes severe line broadening, rendering peak picking and subsequent 1H T2 measurements, at high concentration, error prone. This is relevant for mAb-5. For specific mAb-mAb interactions in case of mAb-4, mAb-3, spectral overlap and broad line widths in 1H 1D NMR donot offer the resolution to identify the peaks/ residues involved in specific PPI, because of line widths (150 kDa, 100 kDa protein). As is seen for the system of mAbs in this study, 2D NMR can be reliably used to differentiate at least two different mechanism (specific vs nonspecific) across the five mAbs. In this regard, 2D NMR can be viewed as a “fit for purpose” HOS characterization tool especially when sample availability is low and minimum disruption in experimental workflow is required. Such a knowledge can be utilized to gauge viscosity behavior in Fc engineered mAbs where Fab-Fc interactions can be modulated. Morever, this can be used in introducing mutations in CDR loop to mitigate high viscosity, without compromising the antigen binding. This method can be potentially applied across high viscous mAbs regardless of isotypes, conformational or colloidal behavior as long as a spectrum is available.
Scope and Validity of NMR Fingerprinting in Addressing Viscosity
The 2D methyl NMR enabled HOS fingerprinting of the system of mAbs used in this study demonstrates the potential utility of the technology in the developability stages and/or pre-formulation stages of mAbs and fusion proteins. Fingerprinting by NMR can be used as potential differentiators to identify a viscosity-prone monoclonal antibody from a given set of mAbs under a platform condition. For this purpose, the HOS fingerprints of a well behaved mAb should be available as a reference. The viscosity prone mAbs can either (a) exhibit extensive line broadening (poor S/N) indicative of non-specific PPI or (b) differential peak broadening/peak shift associated with unique set of residues (peaks). Since NMR is a noninvasive technique, the viscosity-prone mAbs can be subjected to altered solution conditions, potentially sampling the formulation design space to identify optimum pH, to obtain a well dispersed spectrum. This provides a good starting point to initiate the screening of viscosity lowering excipients. Addition of viscosity lowering agent would either (a) increase the signal to noise of the mAb spectrum or (b) result in negative any chemical shift changes indicative of PPI. Thus, 2D NMR based methods provide an avenue for rational screening of excipients. Depending on the type of mAbs and their inherent self-associative propensities, methyl based 2D NMR methods could be generally useful in the concentration range around 30–40 mg/mL. Of course, the macroscopic viscosity of mAbs is relatively similar in this concentration range. At high concentrations (> 100 mg/mL), such fingerprinting may have limited utility due to unfavorable NMR relaxation properties and consequent poor S/N ratios. Thus, unlike other methods like AUC or SAXS (small angle X-ray scattering)/SANS (small-angle neutron scattering), 2D NMR-based fingerprinting cannot conclusively identify morphology or size of the clusters. Furthermore, in the case of viscosity prone mAbs which exhibit specific Fab-Fc interactions, selected residues in the Fab domain can be potentially reengineered to identify a low viscosity variant, without compromising overall structure of the mAb. In case of non-specific Fab2-Fab2 interactions, formulation design space needs to be extensively explored or several point mutation (multiple charge reversal) may be required. Lastly, the consequences of HOS fingerprints that are reflective of self-association may also provide clues to protein aggregation and not just increased solution viscosities. In this regard, these fingerprints may provide important insights into biophysical stability of mAbs in the early stages of development.
Conclusions
In this work, viscosity propensities of several mAbs were assessed by viscosity measurements, DLS analysis, and 2D methyl-HSQC NMR. This study demonstrate the utility of 2D NMR methods to identify a viscosity-prone mAb from a well-behaved mAb by comparing respective HOS fingerprints at various solution conditions. This is particularly relevant in the early stages of development when sample amount is limiting. The impact of intact mAbs and its individual structural domains (Fab and Fc) on viscosity properties were studied by 2D NMR methodologies by assessing the comprehensive changes in the HOS fingerprints. While other methods are available to determine the viscosity of mAb solution, 2D NMR provides an insight into the molecular basis of origin of viscosity. Overall, this methodology can be adopted to identify a suitable candidate under a platform condition or to optimize formulations in the preformulation development stages.
Data Availability Statement
The mAbs used in the study are propriety mAbs of Pfizer. The identity of the molecules (sequences) of the mAbs and the raw data are not available publicly. Some of the mAbs are part of the study Pharmaceutical Research volume 31, pages3161–3178 (2014). [and its supplementary information files].
References
Aggarwal SR. What’s fueling the biotech engine—2012 to 2013. Nat Biotechnol. 2014;32(1):32.
Kaplon H, Reichert JM. Antibodies to watch in 2019. MAbs. 2019;11(2):219–38.
Brennan FR, Morton LD, Spindeldreher S, Kiessling A, Allenspach R, Hey A, ... , Sims J. Safety and immunotoxicity assessment of immunomodulatory monoclonal antibodies. MAbs. 2010;2(3):233–255.
Iuga AO, McGuire MJ. Adherence and health care costs. Risk Manag Healthc Policy. 2014;7:35.
Garidel P, Kuhn AB, Schäfer LV, Karow-Zwick AR, Blech M. High-concentration protein formulations: How high is high? Eur J Pharm Biopharm. 2017;119:353–60.
Shieu W, Torhan SA, Chan E, Hubbard A, Gikanga B, Stauch OB, Maa YF. Filling of high-concentration monoclonal antibody formulations into pre-filled syringes: filling parameter investigation and optimization. PDA J Pharm Sci Technol. 2014;68(2):153–63.
Hartl J, Peschel A, Johannsmann D, Garidel P. Characterizing protein–protein-interaction in high-concentration monoclonal antibody systems with the quartz crystal microbalance. Phys Chem Chem Phys. 2017;19(48):32698–707.
Hung JJ, Dear BJ, Karouta CA, Chowdhury AA, Godfrin PD, Bollinger JA, ... , Sharma A. Protein–Protein Interactions of Highly Concentrated Monoclonal Antibody Solutions via Static Light Scattering and Influence on the Viscosity. J Phys Chem B. 2019;123(4):739–755.
Liu J, Nguyen MD, Andya JD, Shire SJ. Reversible self-association increases the viscosity of a concentrated monoclonal antibody in aqueous solution. J Pharm Sci. 2005;94(9):1928–40.
Yearley EJ, Zarraga IE, Shire SJ, Scherer TM, Gokarn Y, Wagner NJ, Liu Y. Small-angle neutron scattering characterization of monoclonal antibody conformations and interactions at high concentrations. Biophys J. 2013;105(3):720–31.
Yadav S, Laue TM, Kalonia DS, Singh SN, Shire SJ. The influence of charge distribution on self-association and viscosity behavior of monoclonal antibody solutions. Mol Pharm. 2012;9(4):791–802.
Arora J, Hu Y, Esfandiary R, Sathish HA, Bishop SM, Joshi SB., ... , Weis DD. Charge-mediated Fab-Fc interactions in an IgG1 antibody induce reversible self-association, cluster formation, and elevated viscosity. MAbs. 2016;8(8):1551–1574.
Yadav S, Shire SJ, Kalonia DS. Viscosity behavior of high-concentration monoclonal antibody solutions: correlation with interaction parameter and electroviscous effects. J Pharm Sci. 2012;101(3):998–1011.
Yadav S, Shire SJ, Kalonia DS. Factors affecting the viscosity in high concentration solutions of different monoclonal antibodies. J Pharm Sci. 2010;99(12):4812–29.
Sule SV, Cheung JK, Antochshuk V, Bhalla AS, Narasimhan C, Blaisdell S, ... , Tessier PM. Solution pH that minimizes self-association of three monoclonal antibodies is strongly dependent on ionic strength. Mol Pharm 2012;9(4):744–751.
Yadav S, Sreedhara A, Kanai S, Liu J, Lien S, Lowman H, ... , Shire SJ. Establishing a link between amino acid sequences and self-associating and viscoelastic behavior of two closely related monoclonal antibodies. Pharm Res. 2011;28(7):1750–1764.
Lehermayr C, Mahler HC, Mäder K, Fischer S. Assessment of net charge and protein–protein interactions of different monoclonal antibodies. J Pharm Sci. 2011;100(7):2551–62.
Esfandiary R, Parupudi A, Casas-Finet J, Gadre D, Sathish H. Mechanism of reversible self-association of a monoclonal antibody: role of electrostatic and hydrophobic interactions. J Pharm Sci. 2015;104(2):577–86.
Binabaji E, Ma J, Zydney AL. Intermolecular interactions and the viscosity of highly concentrated monoclonal antibody solutions. Pharm Res. 2015;32(9):3102–9.
Neergaard MS, Kalonia DS, Parshad H, Nielsen AD, Møller EH, van de Weert M. Viscosity of high concentration protein formulations of monoclonal antibodies of the IgG1 and IgG4 subclass–Prediction of viscosity through protein–protein interaction measurements. Eur J Pharm Sci. 2013;49(3):400–10.
Connolly BD, Petry C, Yadav S, Demeule B, Ciaccio N, Moore JM, ... , Gokarn, Y. R., Weak interactions govern the viscosity of concentrated antibody solutions: high-throughput analysis using the diffusion interaction parameter. Biophys J. 2012;103(1):69–78.
Geng SB, Wittekind M, Vigil A, Tessier PM. Measurements of monoclonal antibody self-association are correlated with complex biophysical properties. Mol Pharm. 2016;13(5):1636–45.
Liu Y, Caffry I, Wu J, Geng SB, Jain T, Sun T, ... , Vásquez M. High-throughput screening for developability during early-stage antibody discovery using self-interaction nanoparticle spectroscopy. MAbs. 2014;6(2):483–492.
Saito S, Hasegawa J, Kobayashi N, Kishi N, Uchiyama S, Fukui K. Behavior of monoclonal antibodies: relation between the second virial coefficient (B 2) at low concentrations and aggregation propensity and viscosity at high concentrations. Pharm Res. 2012;29(2):397–410.
Scherer TM, Liu J, Shire SJ, Minton AP. Intermolecular interactions of IgG1 monoclonal antibodies at high concentrations characterized by light scattering. J Phys Chem B. 2010;114(40):12948–57.
Godfrin PD, Zarraga IE, Zarzar J, Porcar L, Falus P, Wagner NJ, Liu Y. Effect of hierarchical cluster formation on the viscosity of concentrated monoclonal antibody formulations studied by neutron scattering. J Phys Chem B. 2016;120(2):278–91.
Fukuda M, Moriyama C, Yamazaki T, Imaeda Y, Koga A. Quantitative correlation between viscosity of concentrated MAb solutions and particle size parameters obtained from small-angle x-ray scattering. Pharm Res. 2015;32(12):3803–12.
Woldeyes MA, Qi W, Razinkov VI, Furst EM, Roberts CJ. How well do low-and high-concentration protein interactions predict solution viscosities of monoclonal antibodies? J Pharm Sci. 2019;108(1):142–54.
Ramallo N, Paudel S, Schmit J. The Journal of Physical Chemistry B. 2019;123(18):3916–23. https://doi.org/10.1021/acs.jpcb.9b01511.
von Bülow S, Siggel M, Linke M, Hummer G. Dynamic cluster formation determines viscosity and diffusion in dense protein solutions. Proc Natl Acad Sci U S A. 2019;116(20):9843–52. https://doi.org/10.1073/pnas.1817564116.
Allmendinger A, Fischer S, Huwyler J, Mahler HC, Schwarb E, Zarraga IE, Mueller R. Rheological characterization and injection forces of concentrated protein formulations: an alternative predictive model for non-Newtonian solutions. Eur J Pharm Biopharm. 2014;87(2):318–28.
Li X, Geng SB, Chiu ML, Saro D, Tessier PM. High-throughput assay for measuring monoclonal antibody self-association and aggregation in serum. Bioconjug Chem. 2015;26(3):520–8.
Dear BJ, et al. Enhancing stability and reducing viscosity of a monoclonal antibody with cosolutes by weakening protein-protein interactions. J Pharm Sci. 2019;108.8:2517–26.
Borwankar AU, Dear BJ, Twu A, Hung JJ, Dinin AK, Wilson BK, ... , Johnston KP. Viscosity reduction of a concentrated monoclonal antibody with arginine· HCl and arginine· glutamate. Ind Eng Chem Res. 2016;55(43):11225–11234.
Qun D, et al. Process optimization and protein engineering mitigated manufacturing challenges of a monoclonal antibody with liquid-liquid phase separation issue by disrupting inter-molecule electrostatic interactions. MAbs. 2019;11(4). Taylor & Francis.
Geng SB, Cheung JK, Narasimhan C, Shameem M, Tessier PM. Improving monoclonal antibody selection and engineering using measurements of colloidal protein interactions. J Pharm Sci. 2014;103(11):3356–63.
Geoghegan JC, Fleming R, Damschroder M, Bishop SM, Sathish HA, Esfandiary R. Mitigation of reversible self-association and viscosity in a human IgG1 monoclonal antibody by rational, structure-guided Fv engineering. MAbs. 2016;8(5):941–50.
Chow C-K, Allan BW, Chai Q, Atwell S, Lu J. Therapeutic antibody engineering to improve viscosity and phase separation guided by crystal structure. Mol Pharm. 2016;13(3):915–23.
Guo Z, Chen A, Nassar RA, Helk B, Mueller C, Tang Y, ... , Klibanov AM. Structure-activity relationship for hydrophobic salts as viscosity-lowering excipients for concentrated solutions of monoclonal antibodies. Pharm Res. 2012;29(11):3102–3109.
Hung JJ, Dear BJ, Dinin AK, Borwankar AU, Mehta SK, Truskett TT, Johnston KP. Improving viscosity and stability of a highly concentrated monoclonal antibody solution with concentrated proline. Pharm Res. 2018;35(7):133.
Ke P, Batalha IL, Dobson A, Tejeda-Montes E, Ekizoglou S, Christie G, ... , van der Walle CF. Novel salts of dipicolinic acid as viscosity modifiers for high concentration antibody solutions. Int J Pharm. 2018;548(1):682–688.
Poppe L, Jordan JB, Lawson K, Jerums M, Apostol I, Schnier PD. Profiling formulated monoclonal antibodies by 1H NMR spectroscopy. Anal Chem. 2013;85(20):9623–9.
Tomar DS, Li L, Broulidakis MP, Luksha NG, Burns CT, Singh SK, Kumar S. In-silico prediction of concentration-dependent viscosity curves for monoclonal antibody solutions. MAbs. 2017;9(3):476–89.
Kanai S, Liu JUN, Patapoff TW, Shire SJ. Reversible self-association of a concentrated monoclonal antibody solution mediated by Fab-Fab interaction that impacts solution viscosity. J Pharm Sci. 2008;97(10):4219–27.
Arora J, Hickey JM, Majumdar R, Esfandiary R, Bishop SM, Samra HS, ... , Volkin DB. Hydrogen exchange mass spectrometry reveals protein interfaces and distant dynamic coupling effects during the reversible self-association of an IgG1 monoclonal antibody. MAbs. 2015;7(3):525–539.
Majumdar R, Middaugh CR, Weis DD, Volkin DB. Hydrogen-deuterium exchange mass spectrometry as an emerging analytical tool for stabilization and formulation development of therapeutic monoclonal antibodies. J Pharm Sci. 2015;104(2):327–45.
Li L, Kumar S, Buck PM, Burns C, Lavoie J, Singh SK, Warne NW, Nichols P, Luksha N, Boardman D. Concentration dependent viscosity of monoclonal antibody solutions: explaining experimental behavior in terms of molecular properties. Pharm Res. 2014;31(11):3161–78.
Majumder S, Saati A, Philip S, Liu LL, Stephens E, Rouse JC, Alphonse Ignatius A. Utility of High Resolution NMR Methods to Probe the Impact of Chemical Modifications on Higher Order Structure of Monoclonal Antibodies in Relation to Antigen Binding. Pharm Res. 2019;36(9):130.
Majumder S, Jones MT, Kimmel M, Alphonse Ignatius A. Probing Conformational Diversity of Fc Domains in Aggregation-Prone Monoclonal Antibodies. Pharm Res. 2018;35(11):220.
Arbogast LW, Delaglio F, Schiel JE, Marino JP. Multivariate analysis of two-dimensional 1H, 13C methyl NMR spectra of monoclonal antibody therapeutics to facilitate assessment of higher order structure. Anal Chem. 2017;89(21):11839–45.
Bhattacharya D, et al. Conformational study on pheromonotropin neuropeptide using NMR and molecular dynamics. Pharm Anal Acta. 2015;6.359.10:4172.
Guan X, Zhang L, Wypych J. Direct mass spectrometric characterization of disulfide linkages. MAbs. 2018;10(4):572–82.
Pyae P, et al. Understanding molecular mechanisms of biologics drug delivery and stability from NMR spectroscopy. Adv Drug Deliv Rev. 2021;174:1–29.
Majumder S, Wang W, Ignatius AA. Impact of buffers on colloidal property and aggregation propensities of a bispecific antibody. J Pharm Sci. 2019;108(3):1139–47.
Dear BJ, Chowdhury A, Hung JJ, Karouta CA, Ramachandran K, Nieto MP, ... , Johnston KP. Relating collective diffusion, protein–protein interactions, and viscosity of highly concentrated monoclonal antibodies through dynamic light scattering. Ind Eng Chem Res. 2019;58(50):22456–71.
ACKNOWLEDGMENTS AND DISCLOSURES
The authors acknowledge Dr. Markus Voehler in scheduling and assisting with NMR experiments at the high-resolution Biomolecular NMR facility at Vanderbilt University. Sincere thanks to Bob Walters, Victor Beaumont, Kelly Sackett and Hai-Young Kim for the critical review of the manuscript. The authors also thank Bob Walters and Anjali Mehta for generating selected viscosity profiles of mAbs. All authors are either current or ex-employees of Pfizer.
Funding
A postdoctoral fellowship to S.M. by Pfizer Inc. is gratefully acknowledged.
Author information
Authors and Affiliations
Contributions
SM, AAI conceived the project. SM and DSB performed NMR experiments, data analysis and wrote the manuscript. SM, AL, performed glycerol based viscosity experiments. SM, AL, AAI, DSB performed biophysical experiments. All authors contributed to improving the manuscript.
Corresponding author
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Majumder, S., Bhattacharya, D.S., Langford, A. et al. Utility of High Resolution 2D NMR Fingerprinting in Assessing Viscosity of Therapeutic Monoclonal Antibodies. Pharm Res 39, 529–539 (2022). https://doi.org/10.1007/s11095-022-03200-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11095-022-03200-6